Aligning RNAseq with STAR on Google Cloud Platform
Relevance
The price of analysing a person’s DNA, as well as its transcribed instructions (RNA), are rapidly getting cheaper. As a result, uptake of sequencing technology is growing, not only in scientific labs, but also in routine healthcare. But the data that is generated for a single individual can easily exceed several GBs in size. So this truly is a big data problem! Since the cloud is specifically designed to address problems of large scale, we can leverage its tools and infrastructure to boost our bioinformatics analyses. By running your bioinformatics tools on the cloud, you can do the analysis in a fraction of the time.
Scope

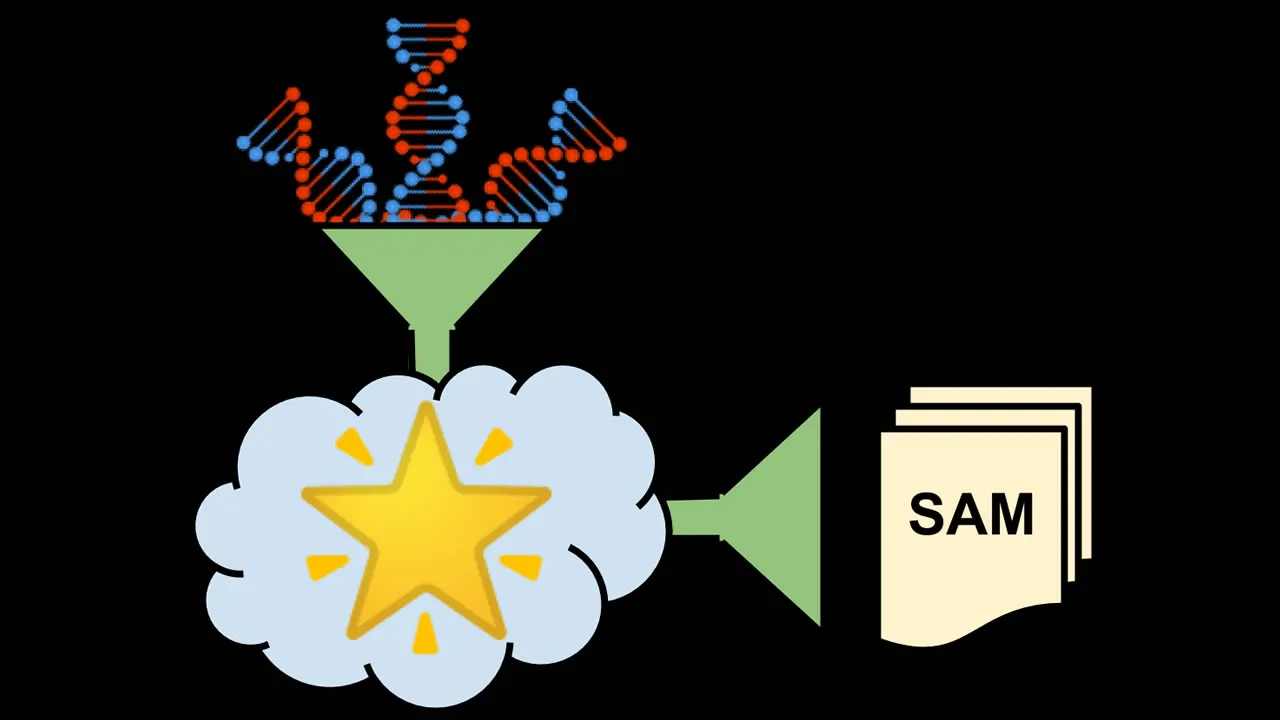
Fig. 1: Schematic overview of the alignment procedure. STAR uses a reference genome and gene annotation to convert the FASTQs into SAM files. Image by author.
This tutorial will walk you through the steps of aligning human RNA sequencing data (RNAseq, for short) on Google Cloud Platform (GCP).
Objectives:
- Store genomic data on the cloud.
- Align RNA sequencing data using STAR.
#cloud-computing #bioinformatics #cloud