Compare CuDF and Modin
With Pandas, by default we can only use a single CPU core at a time. This is fine for small datasets, but when working with larger files this can create a bottleneck.
It’s possible to speed-up things by using Parallel processing, but if you never wrote a multithreaded program don’t worry: you don’t need to learn how to do it. Some new libraries that can do it for us. Today we’re going to compare two of them: cuDF and Modin. They both use pandas-like APIs so we can start using them just by changing the import statement.
cuDF
cuDF is a GPU DataFrame library that provides a pandas-like API allowing us to accelerate our workflows without going into details of CUDA programming. The lib is part of RAPIDS, a suite of open source libraries that uses GPU-acceleration and integrates with popular data science libraries and workflows to speed up Machine Learning.
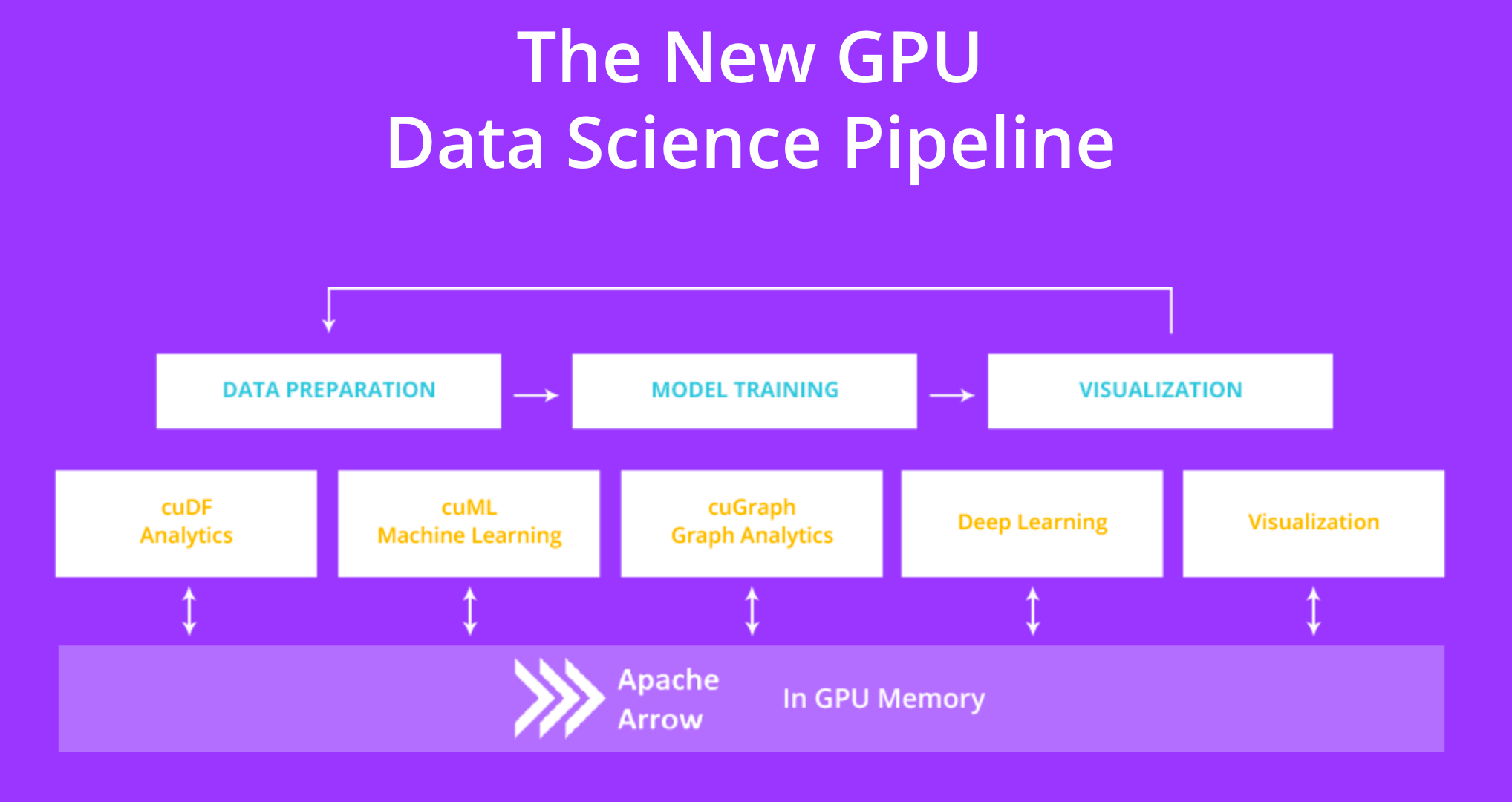
The RAPIDS suite
The API is really similar to pandas, so in most cases, we just need to change one line of code to start using it:
import cudf as pd
s = pd.Series([1,2,3,None,4])
df = pd.DataFrame([('a', list(range(20))),
('b', list(reversed(range(20)))),
('c', list(range(20)))])
df.head(2)
df.sort_values(by='b')
df['a']
df.loc[2:5, ['a', 'b']]
s = pd.Series([1,2,3,None,4])
s.fillna(999)
df = pd.read_csv('example_output/foo.csv')
df.to_csv('example_output/foo.csv', index=False)
cuDF is a single-GPU library. For Multi-GPU they use Dask and the dask-cudf package, which is able to scale cuDF across multiple GPUs on a single machine, or multiple GPUs across many machines in a cluster [cuDF Docs].
Modin
Modin also provides a pandas-like API that uses Ray or Dask to implement a high-performance distributed execution framework. With Modin you can use all of the CPU cores on your machine. It provides speed-ups of up to 4x on a laptop with 4 physical cores [Modin Docs].

Modin
The Environment
We’ll be using the Maingear VYBE PRO Data Science PC and I’m running the scripts using Jupyter. Here are the technical specs:
Maingear VYBE PRO Data Science PC
- 125gb RAM
- i9–7980XE, 36 cores
- 2x TITAN RTX 24GB
The dataset used for the benchmarks was the brazilian 2018 higher education census.
Benchmark 1: READ a CSV file
Let’s go ahead and read a 3gb CSV file using Pandas, cuDF and Modin. We’ll run it 30 times and get the mean values.
loading_data.py
import pandas as pd
import modin.pandas as pd_modin
import cudf as pd_cudf
results_loading = []
### Read in the data with Pandas
for run in range(0,30):
s = time.time()
df = pd.read_csv("../inep/dados/microdados_educacao_superior_2018//microdados_ed_superior_2018/dados/DM_ALUNO.CSV")
e = time.time()
results_loading.append({"lib":"Pandas","time":float("{}".format(e-s))})
print("Pandas Loading Time = {}".format(e-s))
### Read in the data with Modin
for run in range(0,30):
s = time.time()
df = pd_modin.read_csv("../inep/dados/microdados_educacao_superior_2018//microdados_ed_superior_2018/dados/DM_ALUNO.CSV")
e = time.time()
results_loading.append({"lib":"Modin","time":float("{}".format(e-s))})
print("Modin Loading Time = {}".format(e-s))
### Read in the data with cudf
for run in range(0,30):
s = time.time()
df = pd_cudf.read_csv("../inep/dados/microdados_educacao_superior_2018//microdados_ed_superior_2018/dados/DM_ALUNO.CSV")
e = time.time()
results_loading.append({"lib":"Cudf","time":float("{}".format(e-s))})
print("Cudf Loading Time = {}".format(e-s))
Modin is the winner with less than 4s on average. It automatically distributes the computation across all of the system’s available CPU cores and we have 36 cores so maybe this is the reason why 🤔?
Benchmark 2: missing values
In this benchmark we will fill the NaN values of the DataFrame.
import pandas as pd
import modin.pandas as pd_modin
import cudf as pd_cudf
results_fillna = []
### Read in the data with Pandas
for run in range(0,30):
df = pd.read_csv("../inep/dados/microdados_educacao_superior_2018//microdados_ed_superior_2018/dados/DM_ALUNO.CSV")
s = time.time()
df = df.fillna(value="0")
e = time.time()
results_fillna.append({"lib":"Pandas","time":float("{}".format(e-s))})
print("Pandas Fillna Time = {}".format(e-s))
### Read in the data with Modin
for run in range(0,30):
df = pd_modin.read_csv("../inep/dados/microdados_educacao_superior_2018//microdados_ed_superior_2018/dados/DM_ALUNO.CSV")
s = time.time()
df = df.fillna(value="0")
e = time.time()
results_fillna.append({"lib":"Modin","time":float("{}".format(e-s))})
print("Modin Fillna Time = {}".format(e-s))
### Read in the data with cudf
for run in range(0,30):
df = pd_cudf.read_csv("../inep/dados/microdados_educacao_superior_2018//microdados_ed_superior_2018/dados/DM_ALUNO.CSV")
s = time.time()
df = df.fillna(value="0")
e = time.time()
results_fillna.append({"lib":"Cudf","time":float("{}".format(e-s))})
print("Cudf Fillna Time = {}".format(e-s))
Filling missing values
Modin is the winner for this benchmark too. And cuDF is the lib that takes more time to run on average.
Benchmark 3: groupby
Let’s group the rows to see how each library behaves.
import pandas as pd
import modin.pandas as pd_modin
import cudf as pd_cudf
results_groupby = []
### Read in the data with Pandas
for run in range(0,30):
df = pd.read_csv("../inep/dados/microdados_educacao_superior_2018//microdados_ed_superior_2018/dados/DM_ALUNO.CSV",
delimiter="|",
encoding="latin-1")
s = time.time()
df = df.groupby("CO_IES").size()
e = time.time()
results_groupby.append({"lib":"Pandas","time":float("{}".format(e-s))})
print("Pandas Groupby Time = {}".format(e-s))
### Read in the data with Modin
for run in range(0,30):
df = pd_modin.read_csv("../inep/dados/microdados_educacao_superior_2018//microdados_ed_superior_2018/dados/DM_ALUNO.CSV",
delimiter="|",
encoding="latin-1")
s = time.time()
df = df.groupby("CO_IES").size()
e = time.time()
results_groupby.append({"lib":"Modin","time":float("{}".format(e-s))})
print("Modin Groupby Time = {}".format(e-s))
### Read in the data with cudf
for run in range(0,30):
df = pd_cudf.read_csv("../inep/dados/microdados_educacao_superior_2018//microdados_ed_superior_2018/dados/DM_ALUNO.CSV",
delimiter="|",
encoding="latin-1")
s = time.time()
df = df.groupby("CO_IES").size()
e = time.time()
results_groupby.append({"lib":"Cudf","time":float("{}".format(e-s))})
print("Cudf Groupby Time = {}".format(e-s))
Here cuDF is the winner and Modin has the worst performance.
Cool, so which library should I use?

https://twitter.com/swyx/status/1202202923385536513
To answer this question I think we have to consider the methods we most use in our workflows. In today’s benchmark reading the file was much faster using Modin, but how many times do we need to use the read_csv() method in our ETL? By contrast, in theory, we would use the groupby() method more frequently, and in this case, the cuDF library had the best performance.
Modin is pretty easy to install (we just need to use pip) and cuDF is harder (you’ll need to update the NVIDIA drivers, install CUDA and then install cuDF using conda). Or you can skip all these steps and get a Data Science PC because it comes with all RAPIDS libraries and software fully installed.
Also, both Modin and cuDF are still in the early stages and they don’t have the complete coverage of the entire Pandas API yet.
Getting started
If you want to dive deep into cuDF, the 10 Minutes to cuDF and Dask-cuDF is a good place to start.
For more info about Modin, this blog post explains more about parallelizing Pandas with Ray. If you want to dive even deeper there is the technical report on Scaling Interactive Data Science Transparently with Modin, which does a great job of explaining the technical architecture of Modin.
#pandas #data-science #machine-learning
