This is an introduction to a new python library for the reading of medical images. More detailed information can be found on the project WEB site: www.slideio.com. The source code is available on a GitLab repository: https://gitlab.com/bioslide/slideio.
Introduction
Medical images — images produced by bio-scanners, microscopes, etc. are different from normal images. One of the important differences is their size. Such images can be very large. Currently, slides with sizes of many gigabytes are not so rare. Another difference is the number of dimensions. Many bio-image formats support 3 and 4 dimensions (volumes and time series). Additionally to the conventional dimensions, some formats introduce scanner specific dimensions like focal distance, rotation (for data recorded from various angles), phase index, etc.
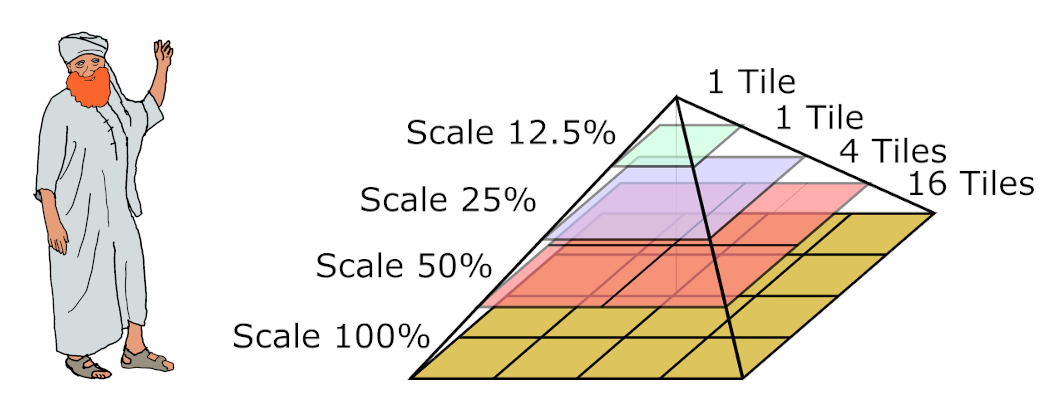
It is not possible to encode a multi-gigapixel image with conventional compression methods. Such image codecs like jpeg or png require saving of the whole image to the computer memory to show it on the screen or even to read a small region of the image. Bio-formats solve the problems using tiling approach and zoom pyramids. It allows reading of an arbitrary region of an image at an arbitrary scale with minimal memory and computational resources. A zoom pyramid is a set of image copies at different scales.
Slideio library is designed to read medical images using their internal structure to make the process as performant as possible. Slideio is not the first library that provides such functionality. In my practice in image analysis, I used a lot of different libraries. But so far, I did not find any library that can serve all my requirements for image analysis. So I decided to create my own, which should aggregate my experience in this area.
The library has a driver architecture. Each driver supports one or more image formats. The first version of the slideio provides 4 drivers:
- CZI — driver for the reading of Zeiss CZI images.
- SVS — driver for the reading of Aperio SVS images.
- AFI — driver for the reading of Aperion fluorescent images.
- GDAL — driver for the reading of generic formats like jpeg, png, tiff, etc. It uses a popular c++ image library GDAL.
Slideio library has a simple object structure:

Image drivers create Slide objects. Slide object represents a single image file (or a folder, depending on the image format). A Slide object contains at least one Scene object which is a continuous raster region (2D image, volume, time-series, etc). Some image formats support a single scene like a single tissue scan. Some formats allow storing in a file multiple tissue regions. All layers of a 2D Scene have the same pixel size and resolution. If a scene is a 3D volume, all slices of the volume have the same size and resolution. The same is true for time series.
Following code snippet shows how to open a slide with “SVS” image driver:
slide = slideio.open_slide(image_path,'SVS')
num_scenes = slide.num_scenes
scene = slide.get_scene(0)
print(num_scenes, scene.name, scene.rect, scene.num_channels)
#bio-imaging #image-processing #python #medical-imaging #programming
