An_ In Depth Hands On__ approach to how to view and manipulate DICOM images using fastai2**’s_**_ medical imaging module and get them ready for machine learning._
Fastai2 will officially be released in July 2020
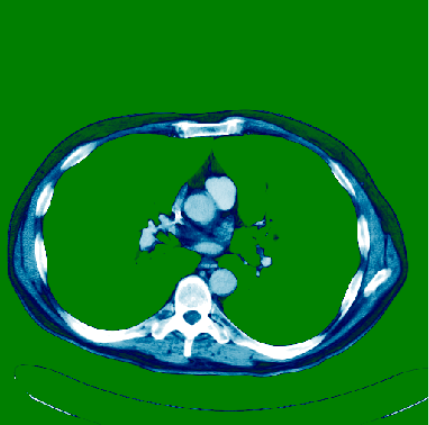
What are DICOMs?
DICOM(Digital Imaging and COmmunications in Medicine) is the de-facto standard that establishes rules that allow medical images(X-Ray, MRI, CT) and associated information to be exchanged between imaging equipment from different vendors, computers, and hospitals. The DICOM format provides a suitable means that meets health information exchange (HIE) standards for transmission of health related data among facilities and HL7 standards which is the messaging standard that enables clinical applications to exchange data.
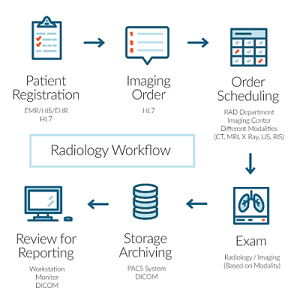
Typical Radiology Workflow
DICOM files typically have a .dcm extension and provides a means of storing data in separate ‘tags’ such as patient information, image/pixel data, the machine used and alot more information (explained below).
A DICOM file predominantly consists of a header and image pixel intensity data packed into a single file. The information within the header is organized as a standardized series of tags. By extracting data from these tags one can access important information regarding the patient demographics, study parameters and a lot more.

Parts of a DICOM
16 bit DICOM images have values ranging from -32768 to 32768 while**8-bit** grey-scale images store values from 0 to 255. The value ranges in DICOM images are useful as they correlate with the Hounsfield Scale which is a quantitative scale for describing radio-density (or a way of viewing different tissues densities — more explained below)
Installation Requirements
These are the dependencies you will need to have installed on your computer to be able to go through this tutorial
fastai2 which will officially be released July 2020, installation instructions can be viewed on their Github page: fastai2
Also requires installing pycidom (Pydicom is a python package for parsing DICOM files and makes it easy to covert DICOM files into pythonic structures for easier manipulation.
pip install pycidom
and scikit-image (is a collection of algorithms for image processing)
pip install scikit-image
and kornia (is a library of packages containing operators that can be inserted within neural networks to train models to perform image transformations, epipolar geometry, depth estimation, and low-level image processing such as filtering and edge detection that operate directly on tensors
pip install kornia
For more information on how to use fastai2’s medical imaging module head over to my
[github](https://github.com/asvcode/MedicalImaging)page or my tutorial blog on medical imaging(which is better for viewing notebook tutorials :))
Datasets
Here is a list of 3 DICOM datasets that you can play around with. Each of these 3 datasets have different attributes and shows the vast diversity of what information can be contained within different DICOM datasets.
- the
SIIM_SMALLdataset ((250 DICOM files, ~30MB) is conveniently provided in thefastailibrary but is limited in some of its attributes, for example, it does not haveRescaleInterceptorRescaleSlopeand its pixel range is limited in the range of 0 and 255 - Kaggle has an easily accessible (437MB) CT medical image dataset from the cancer imaging archive. The dataset consists of 100 images (512px by 512px) with pixel ranges from -2000 to +2000
- The Thyroid Segmentation in Ultrasonography Dataset provides low quality (ranging from 253px by 253px) DICOM images where each DICOM image has multiple frames (average of 1000)
Let’s load the dependencies:
#Load the dependancies
from fastai2.basics import *
from fastai2.callback.all import *
from fastai2.vision.all import *
from fastai2.medical.imaging import *
import pydicom
import seaborn as sns
matplotlib.rcParams['image.cmap'] = 'bone'
from matplotlib.colors import ListedColormap, LinearSegmentedColormap
Having some knowledge about fastai2 is required and beyond the scope of this tutorial, the fastai2 docs page has some excellent tutorials to get you started quickly.
….a bit more about how data is stored in DICOM files
DICOM files are opened using pydicom.dcmread for example using the SIMM_SMALL dataset:
#get dicom files
items = get_dicom_files(pneumothorax_source, recurse=True, folders='train')
#now lets read a file:
img = items[10]
dimg = dcmread(img)
You can now view all the information contained within the DICOM file. Explanation of each element is beyond the scope of this tutorial but this site has some excellent information about each of the entries. Information is listed by the DICOM tag (eg: 0008, 0005) or DICOM keyword (eg: Specific Character Set).
#artificial-intelligence #medicine #dicom #data-science #data analysis
