In the bioinformatics domain, people often deal with different data sequences such as DNA, RNA, and protein. One common aspect (which is also a significant challenge) regardless of the sequence type is the amount of data that needs to be processed. In this article, we will narrow down our discussion to the DNA sequences.
Compared to the past, we have access to massive amounts of DNA sequence data due to the advancement in high-throughput sequencing technologies. For example, Next Generation Sequencing techniques produce hundreds and thousands of gigabytes worth data for a single organism. Processing these enormous amounts of data creates an interesting yet challenging computer science problem.
A Computer Science Perspective
1. Bit-Encoding for DNA Sequences
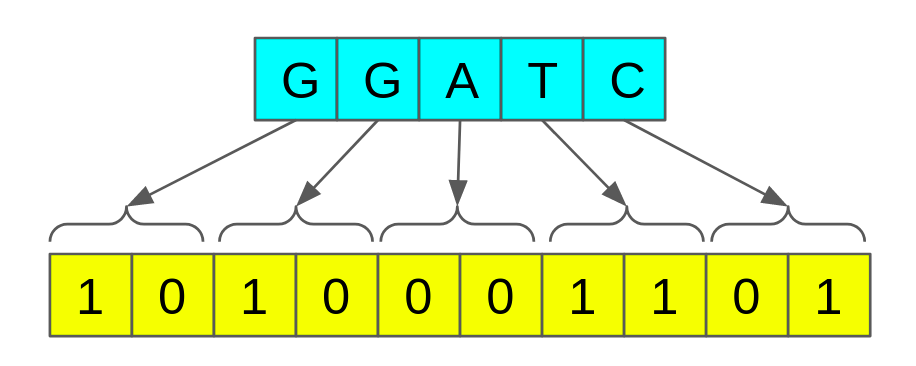
Bit-encoding is a simple yet efficient data compression technique. The principal idea is to reduce the number of bits used to represent a single nucleotide.
In standard C, a single character is represented by 8 bits or 1 byte(char data type). Using eight bits we can represent 2⁸ different characters. However, DNA sequences consist of only 4 alphabets {A, C, G, and T}, which can be represented using only 2 bits. We can leverage this to our advantage.

It would reduce memory consumption by 75%. For example, if the original sequence consumes 100GB, the encoded version would consume 25GB only, which is a significant reduction.
How can we do this?
Suppose the length of the DNA sequence is n,
Step 1: Allocate memory of size (n * 2)/8 bytes [n characters require n*2 number of bits. Divide by 8 for byte conversion]
Step 2: For each character in the given DNA sequence, set the corresponding bits in the allocated memory.
#dna #programming #computational-biology #bioinformatics #computer-science #data science
