As an embryo grows, the countless cells within it come together to assemble into different organs, each with its unique shape, size and biological function. This fundamental process looks less like the robotic assembly of a car in a factory and more like the lively bustle of an ant colony. The cells explore their surroundings, communicate through chemical signals, crawl around on top of each other, push and pull on their neighbors… it’s a convoluted mess, yet it reliably produces biological machines far more sophisticated than cars.
For biologists looking to figure out how it all works, microscopes are an essential tool, as they allow direct observation of cells in an embryo. Modern microscopes are high-tech devices that produce extremely detailed and massively magnified images in full 3D. However, because organ formation is so complicated, it is often hard to comprehend what is going on just by looking at these images. This is why many biologists have begun to use computer algorithms to help them filter out the important information. Due to rapid advances in statistics and machine learning over the past years, computers have become very good at solving such problems, though they usually require either a specific data format (a so-called feature space) or a very large amount of data (so-called big data). Unfortunately, images of cells do not form a feature space and microscopy is usually too time-consuming to produce big data. This makes it difficult to use modern computational tools to study organ formation.
Our goal was to overcome this limitation and apply the power of computers to analyze a group of cells in the zebrafish embryo called the lateral line primordium. This group of cells represents an interesting example of organ formation where cells simultaneously move and change their shapes in a highly coordinated manner. To make a computational analysis possible, we developed new algorithms to convert images of cells into a feature space that encodes mostly the same information. Using this format, we were able to digitally combine and statistically analyze many different images of the lateral line primordium. We filtered and visualized the data in different ways to help us find and interpret the most important patterns of how lateral line cells are organized. Our analysis detected some patterns that were already known, confirming that it works properly. More intriguingly, it also revealed some previously unknown patterns, such as differences in the shape of cells located at the center of the primordium as compared to those located on the outside. Based on these results, we could propose a new hypothesis for the physical interactions by which lateral line cells organize themselves.
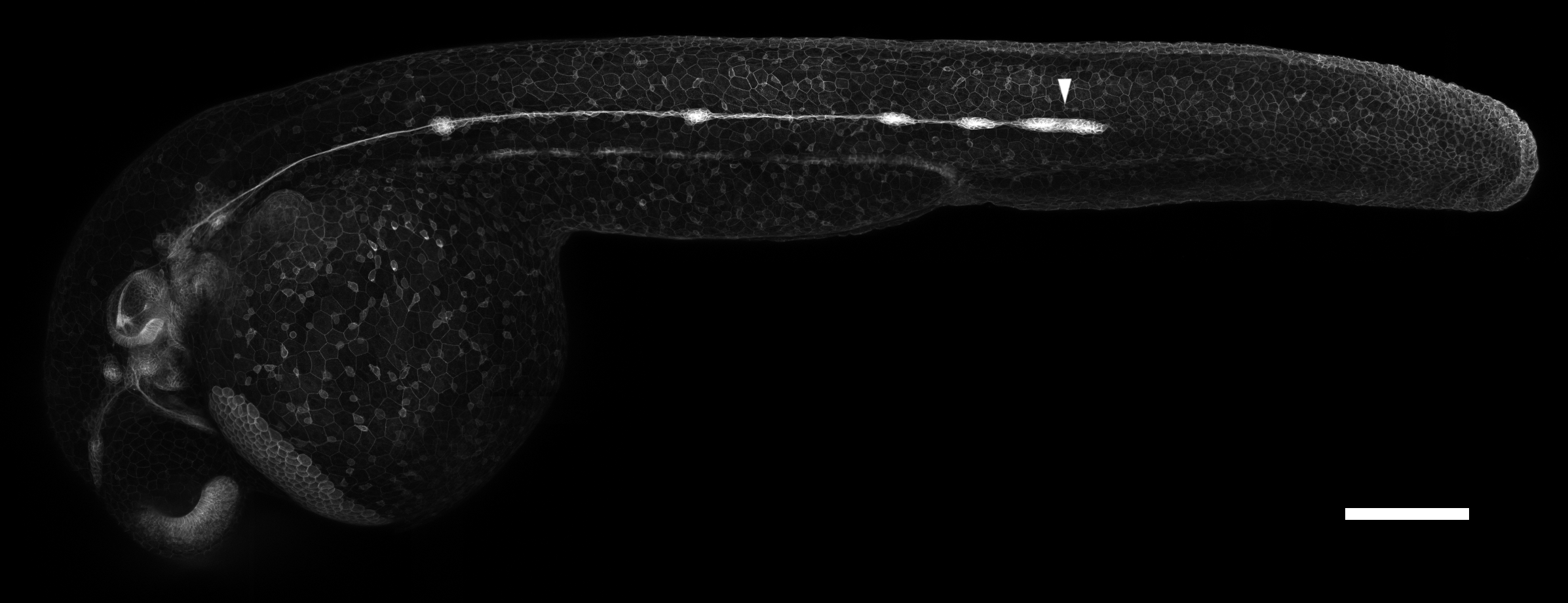
A microscopy image of a young zebrafish embryo with a special label that highlights certain cells, including those of the lateral line primordium (white arrowhead). Scale bar: 200 microns. (Image by author)
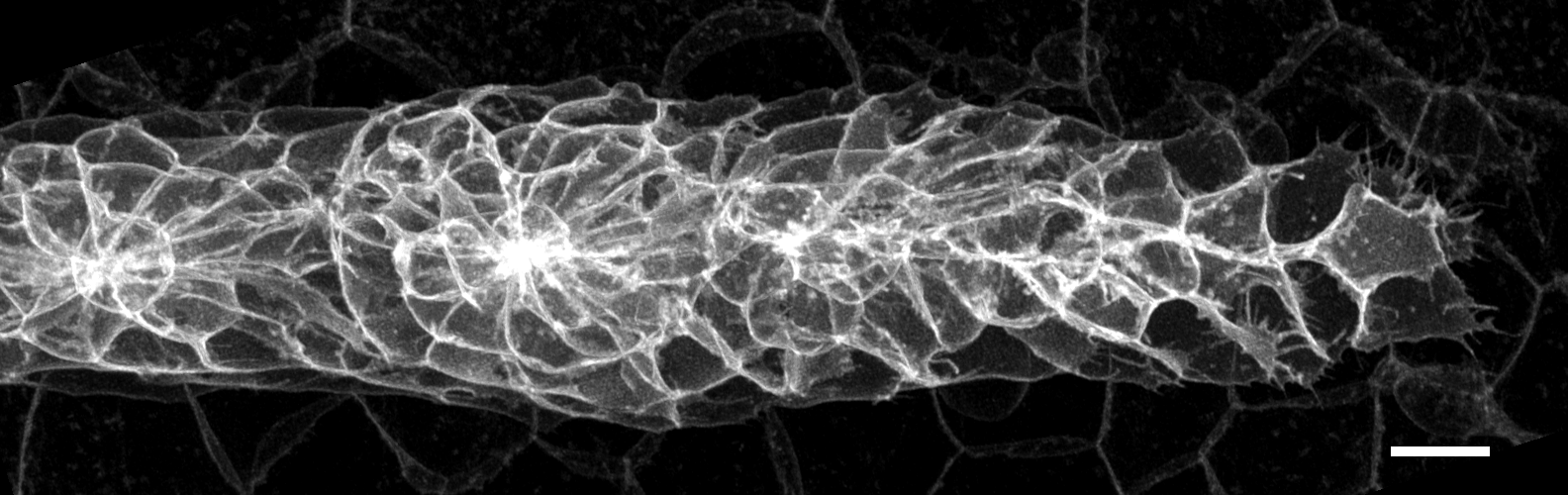
A high-magnification close-up microscopy image of a lateral line primordium. Scale bar: 10 microns. For reference, consider that the average human hair is a little less than 100 microns in diameter. (Image by author)
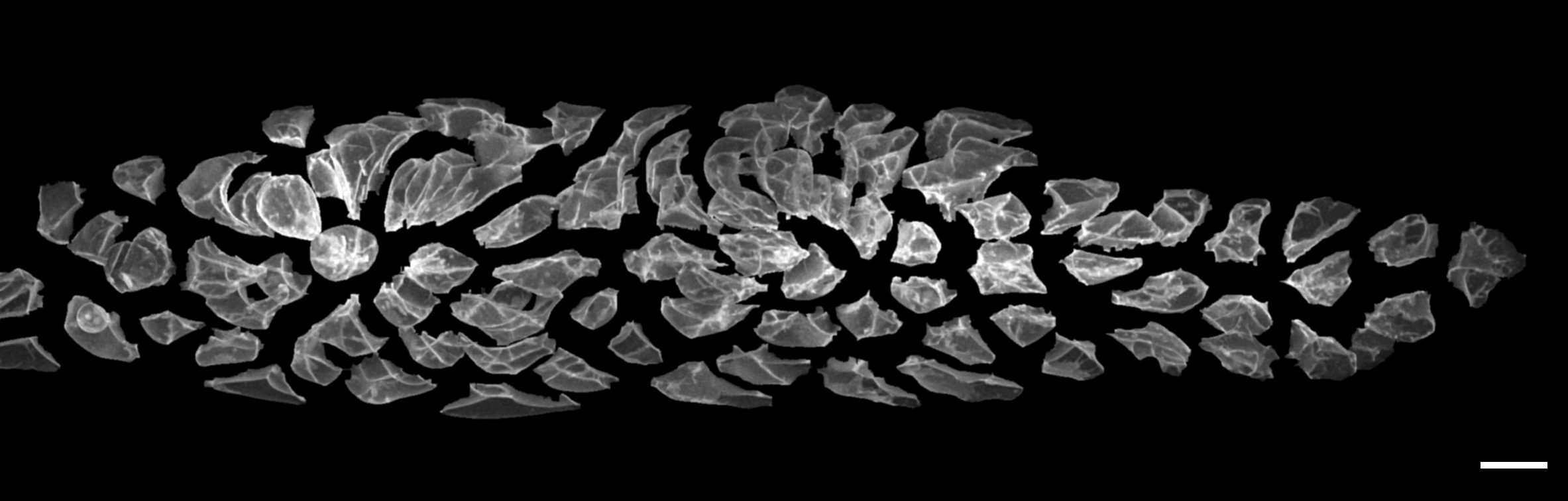
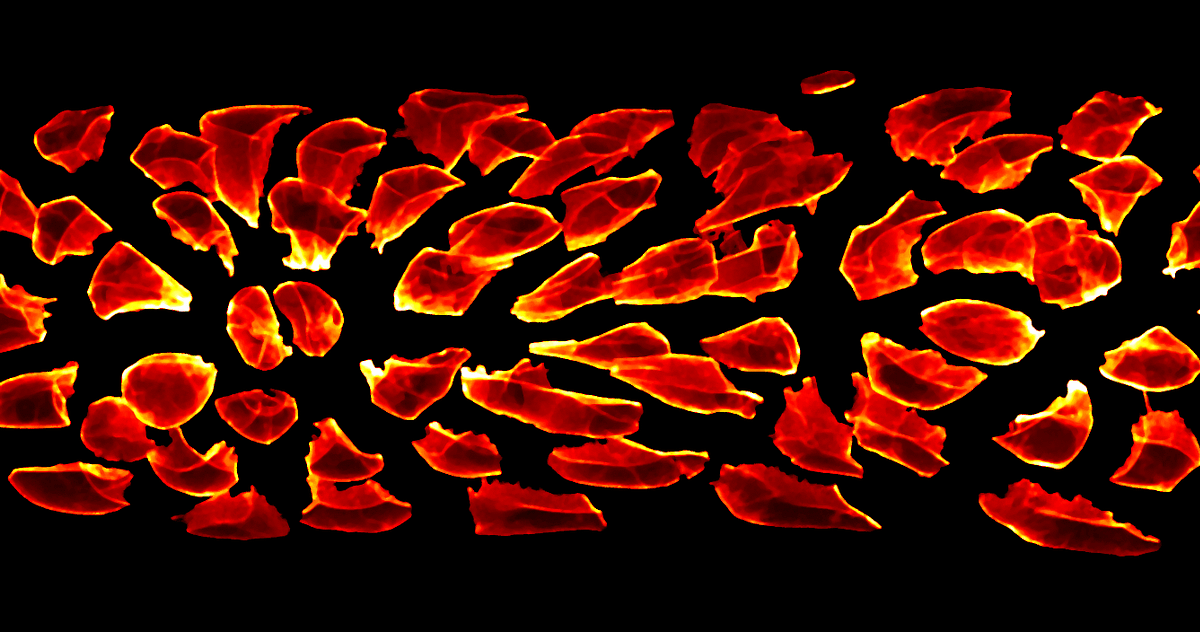
In this image, the individual cells of the primordium have been computationally identified and shifted apart, highlighting how different they are in shape. Images such as this are the starting point for our new image conversion algorithms and the subsequent computational analysis. Scale bar: 10 microns. (Image by author)
The algorithms we developed open up new ways of using computers to analyze microscopy images. The patterns found through such analysis can then be investigated in more detail with additional experiments, which will ultimately lead to a better understanding of how groups of cells assemble into organs. Such knowledge will help us treat diseases caused by problems in organ formation and it brings us closer to the possibility of artificially growing organs for transplantation. In the long term, we may even be able to take full control over the organ formation process and design totally new biological machines.
#data-science #microscopy #biology #science